Overview
Regional Association of Methylome variability with the Exposome and geNome (RAMEN) is an R package whose goal is to identify genome-wide Variable Methylated Loci (VML) from microarray DNA methylation data. Then, using genomic and exposomic data, it can identify which model out of the following explains best the DNA methylation variability at each VML: genetic (G), environmental (E), additive (G+E) or interactive (GxE).
Installation
You can install the development version of RAMEN from GitHub with:
# install.packages("devtools")
devtools::install_github("ErickNavarroD/RAMEN")Core functions
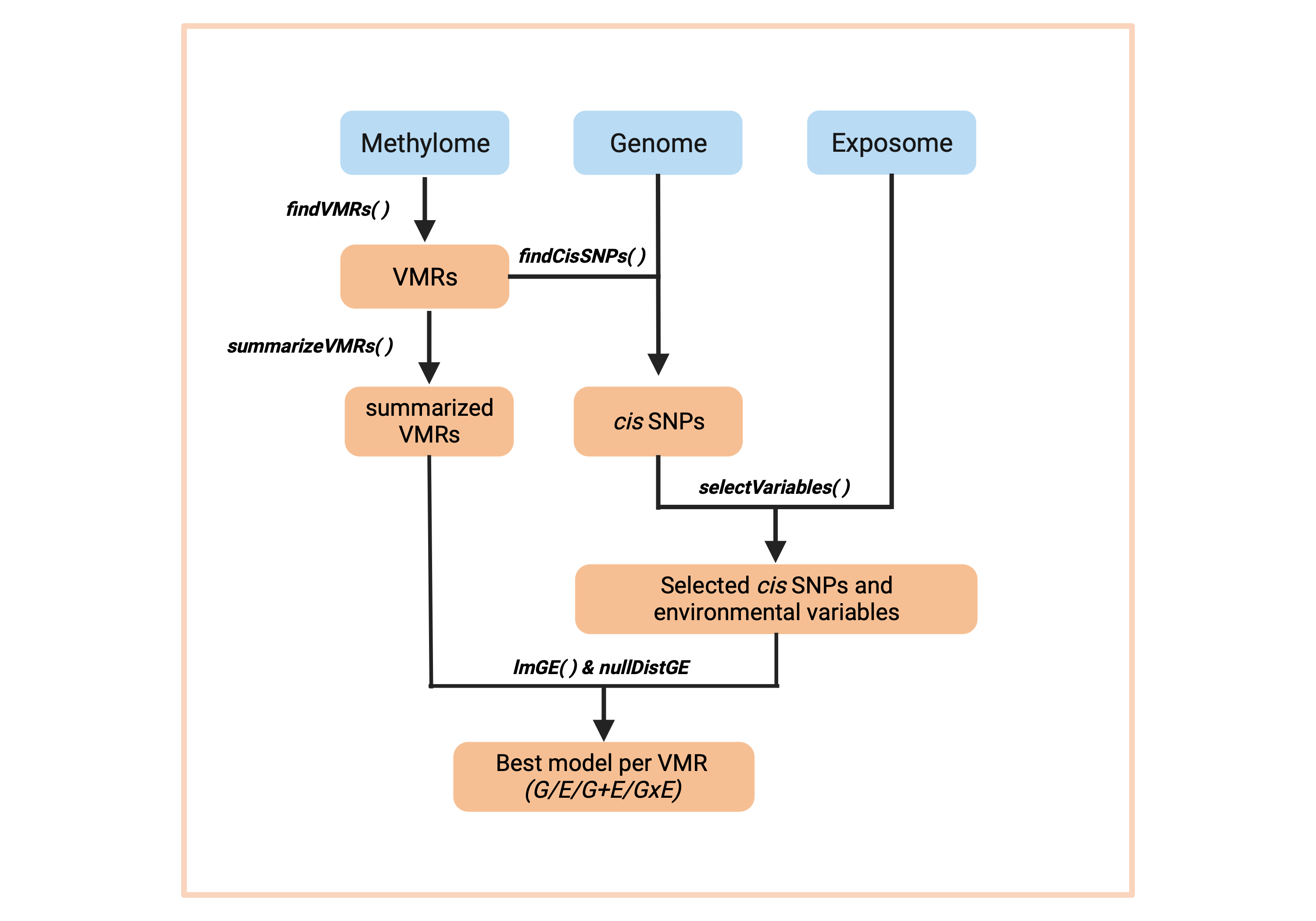
RAMEN consists of six main functions:
-
findVML()identifies Variable Methylated Loci (VML) from microarray data -
summarizeVML()summarizes the regional methylation state of each VML -
findCisSNPs()identifies the SNPs in cis of each VML -
selectVariables()conducts a LASSO-based feature selection strategy to identify potentially relevant cis SNPs and environmental variables -
lmGE()fits linear single-variable genetic (G), environmental (E), pairwise additive (G+E) and pairwise interaction (GxE) linear models, and select the best explanatory model for each VML. -
nullDistGE()simulates a null distribution of G and E effects on DNAme variability. Useful for filtering out poor-performing best explanatory models selected by lmGE().
Altogether, these functions create a pipeline that takes a set of individuals with genome, exposome and DNA methylome information, and generates an estimation of the contribution of genetic variants and environmental exposures to its DNA methylation variability. Functions that conduct computationally intensive tasks are compatible with parallel computing.
Usage
For a detailed tutorial on how to use RAMEN, please check the package’s vignette or website.
Variations to the standard workflow
Besides using RAMEN for a gene-environment contribution analysis, the package provides individual functions that could help users in other tasks, such as:
- Reduction of multiple hypothesis test burden in EWAS or differential methylation analysis by using VML instead of individual probes.
- Fit additive and interaction models given a set of variables of interest and select the best explanatory model for DNAme data (e.g. epistasis or ExE studies).
- Quickly identify SNPs in cis of CpG probes.
- Get the median correlation of probes in custom regions of interest with
medCorVMR().
How to get help for RAMEN
If you have any question about RAMEN usage, please post an issue in this github repository so that future users also benefit from the discussion As an alternative option, you can contact Erick Navarro-Delgado at erick.delgado@ubc.ca.
Acknowledgments
I want to thank Dr. Keegan Korthauer and Dr. Michael S. Kobor for their supervision, feedback and support throughout the development of this package. Also, I want to thank the members of the Kobor and Korthauer lab for their comments and discussion.
The RAMEN package logo was created by Carlos Cortés-Quiñones and Dorothy Lin. Carlos created the draw, and Dorothy refined the logo and did the lettering.
Funding
This work was supported by the University of British Columbia, the BC Children’s Hospital Research Institute and the Social Exposome Cluster.
Citing RAMEN
If you use RAMEN for any of your analyses, please cite the following publication:
- Navarro-Delgado, E.I., Czamara, D., Edwards, K. et al. RAMEN: Dissecting individual, additive and interactive gene-environment contributions to DNA methylome variability in cord blood. Genome Biol 26, 421 (2025). https://doi.org/10.1186/s13059-025-03864-4
Code of conduct
Please note that this package is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.
